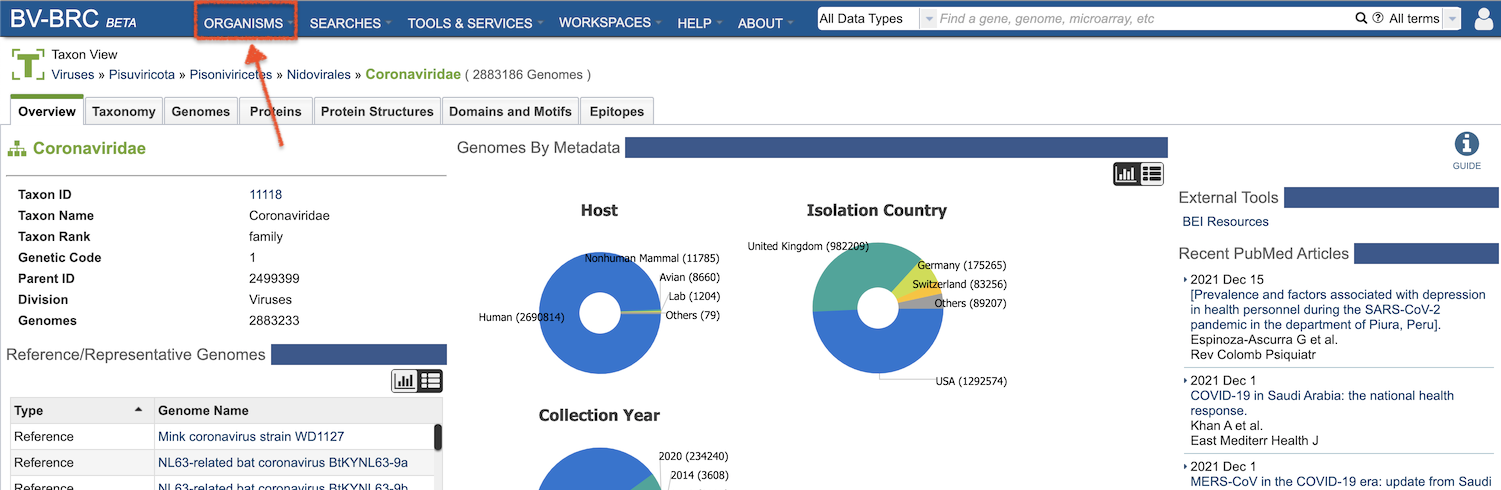
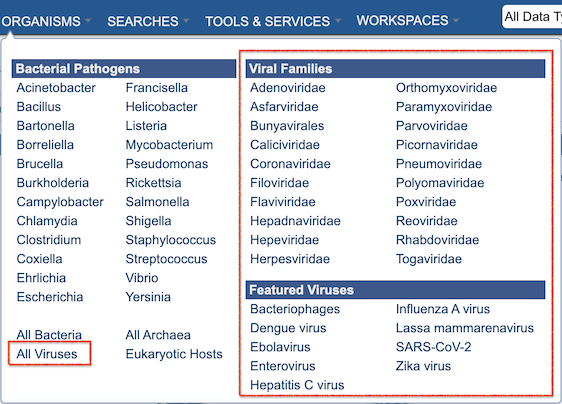
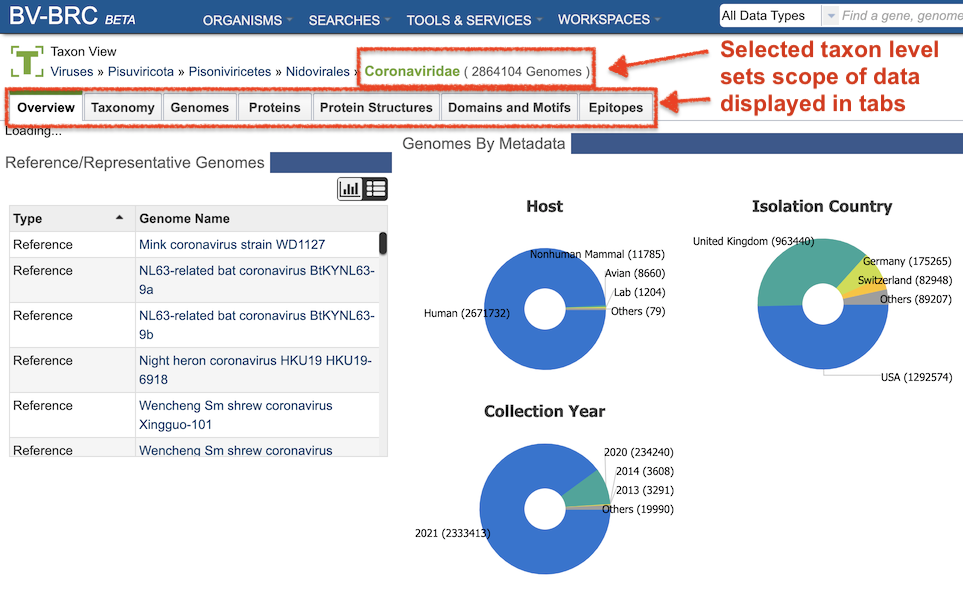
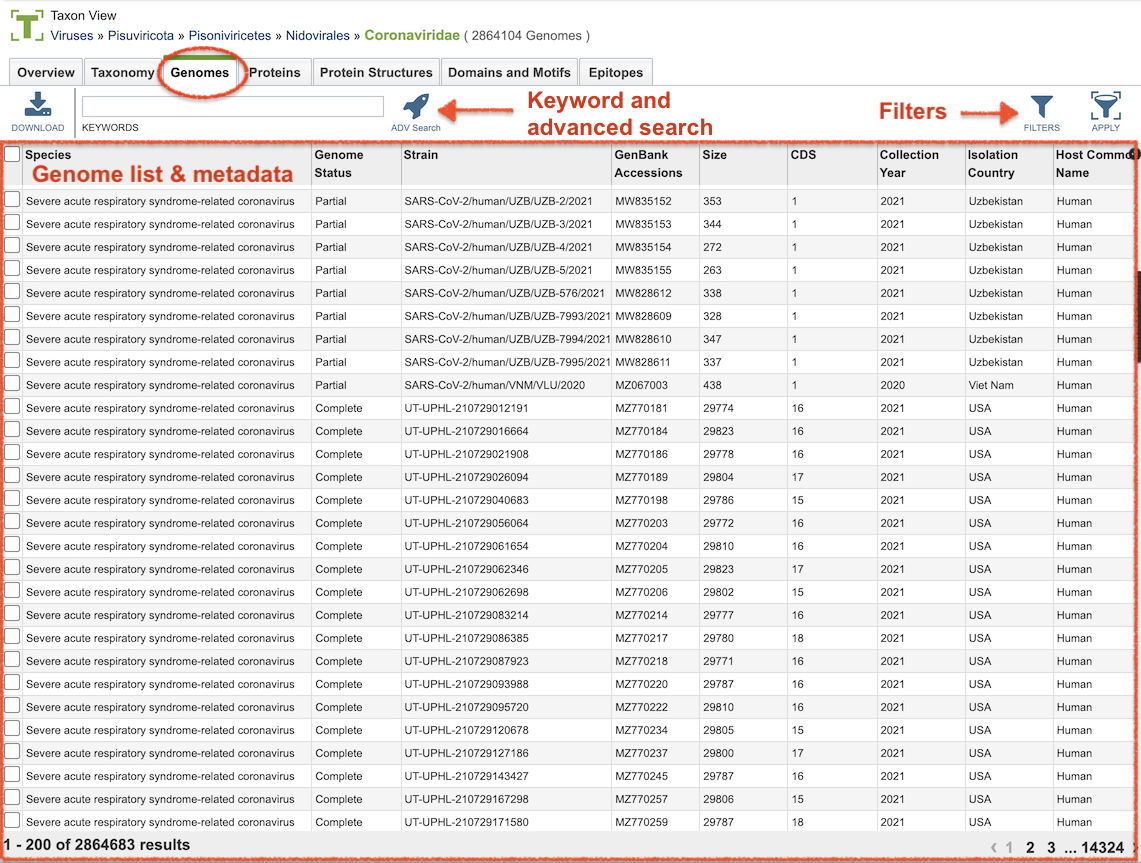
Terminology and Features (for IRD/ViPR Users)¶
BV-BRC integrates key IRD and ViPR viral data and tools with bacterial data and tools from PATRIC. The BV-BRC system is built on the PATRIC system infrastructure (database, services, and website). As a result, the interfaces for accessing and using data and tools differ substantially from IRD and ViPR. To aid researchers in making this transition, the table below provides a mapping of IRD and ViPR features to the corresponding BV-BRC features. Links to relevant BV-BRC help documentation are provided as well.
Additional details are presented below.
Searches¶
Similar to IRD and ViPR, BV-BRC provides searches to facilitate rapide direct access to viral data of interest. Two types of searches are provided: Global Search and Advanced Searches.
Global Search¶
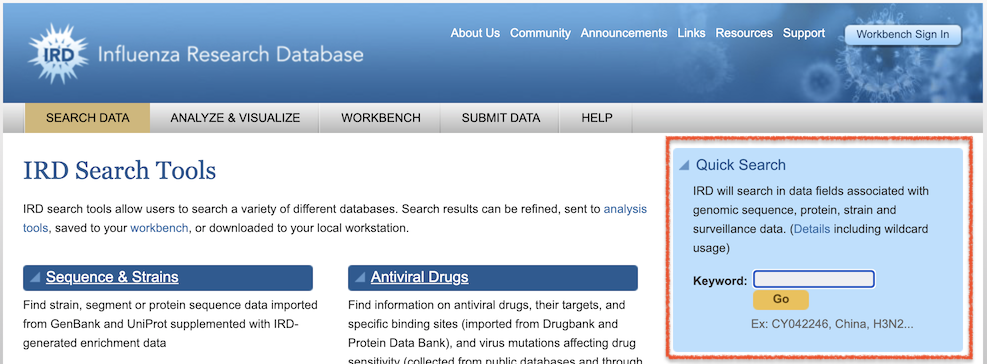
The BV-BRC Global Search provides keyword search capability similar to the IRD/ViPR Quick Search (available from the IRD/ViPR Search Tools page, below), but with advanced controls for selecting data type and other search conditions.
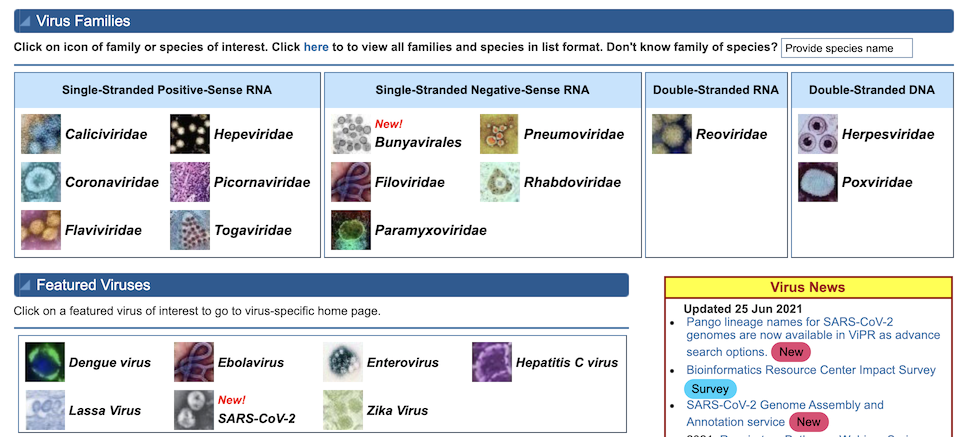
IRD/ViPR Quick Search
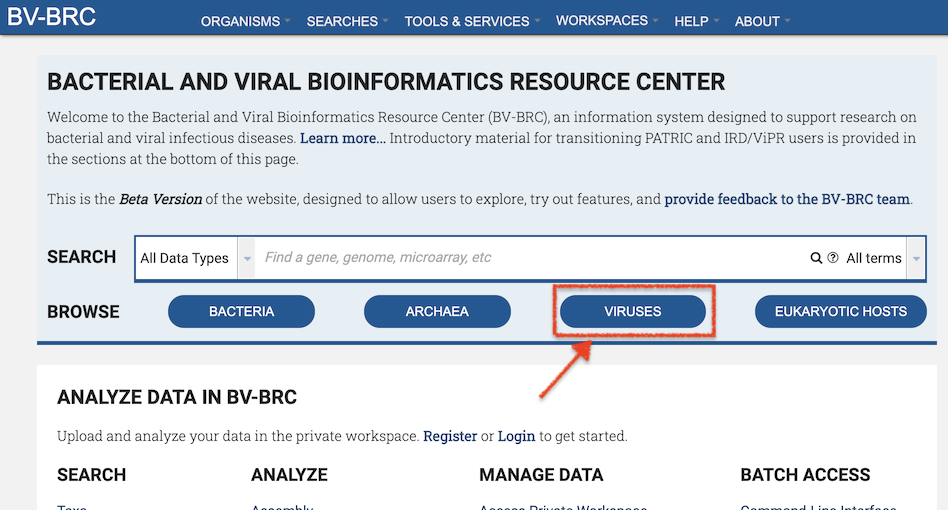
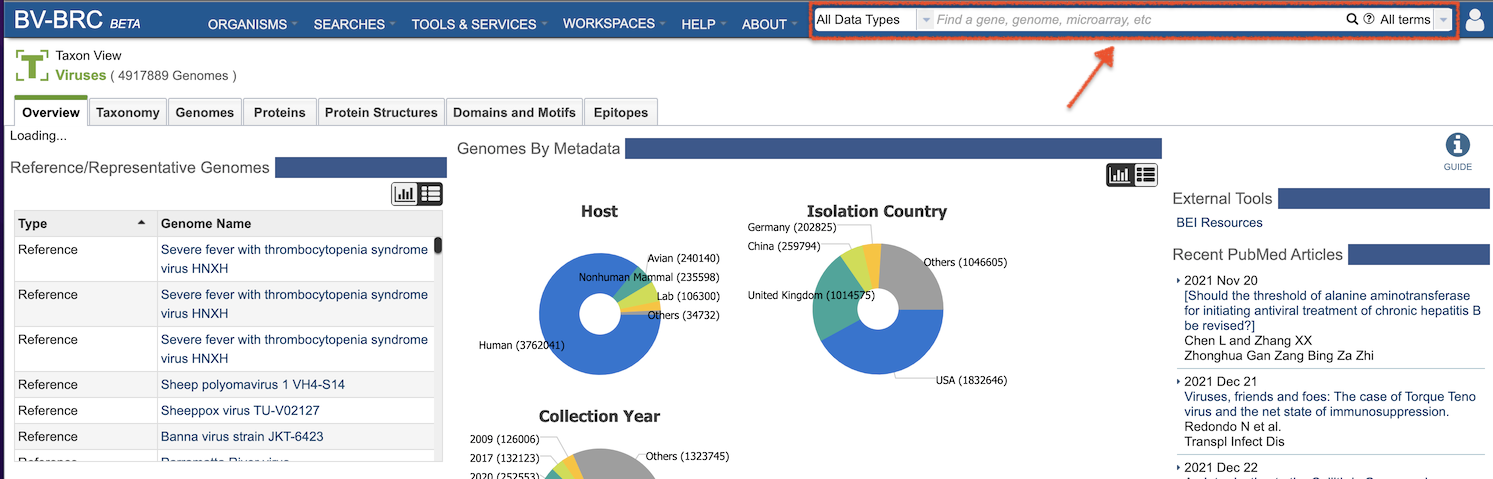
The BV-BRC Global Search is available at the top right of all BV-BRC pages, except for the home page, where it is in the upper center of the page, as shown below.
BV-BRC Global Search
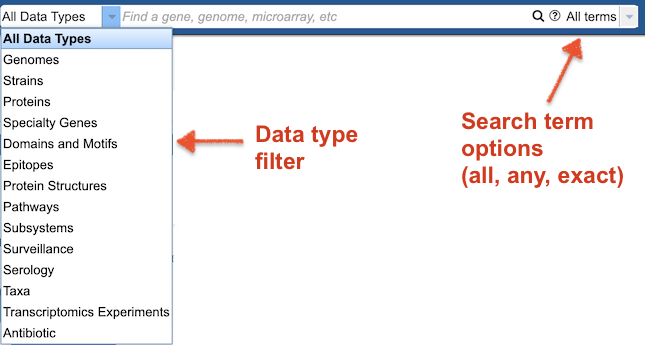
The Global Search provides optional filters for data types and search term options (All, Any, Exact).
Detailed instructions for using the Global Search are available from the BV-BRC Global Search Quick Reference Guide.
Advanced Searches¶
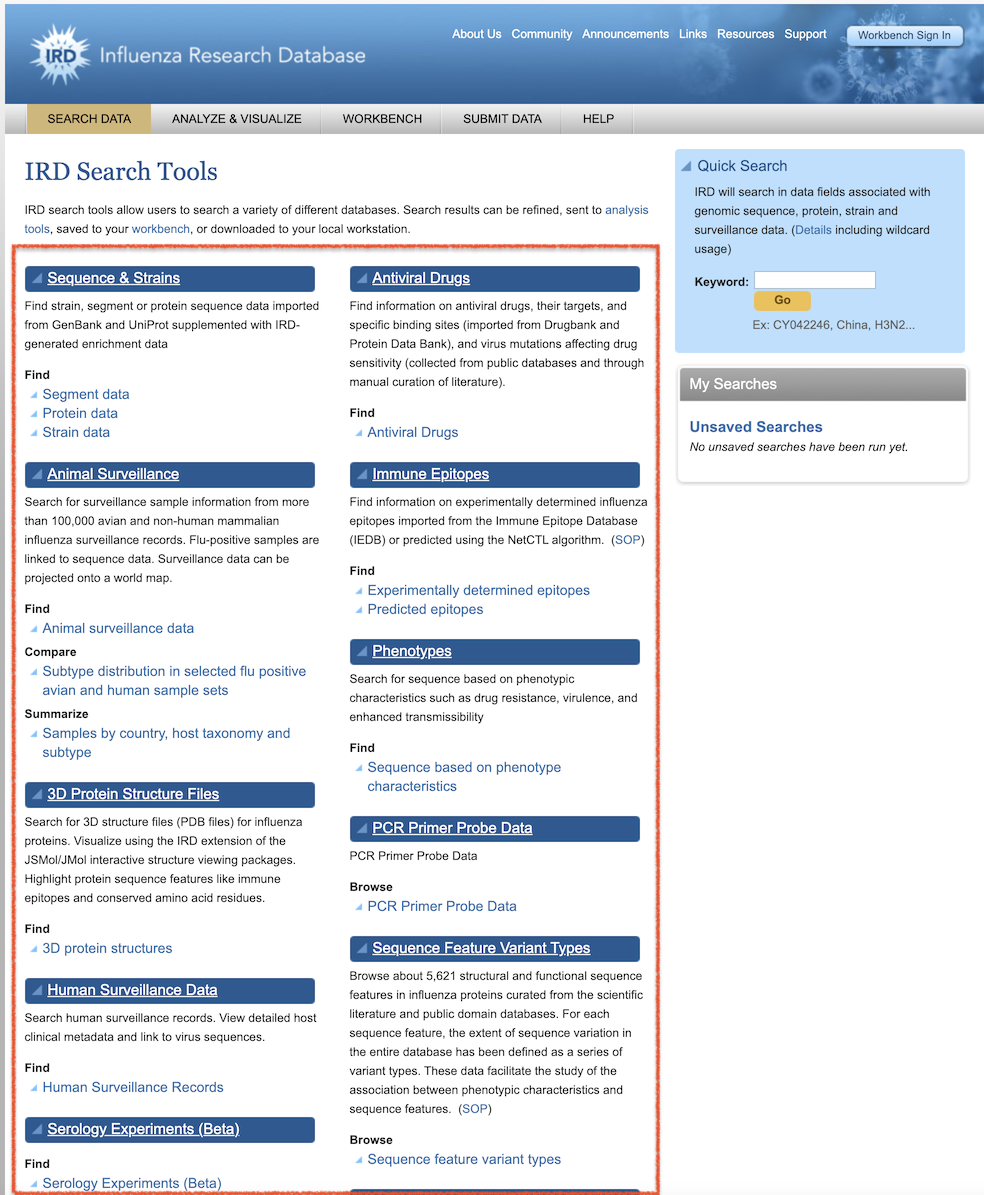
The BV-BRC Advanced Searches provide fine-grained searches based on data type and metadata values, similar to the IRD/ViPR Search Tools (available from the IRD/ViPR Search Tools page, below). Some searches have been deprecated based on limited usage and available data.
IRD/ViPR Search Tools
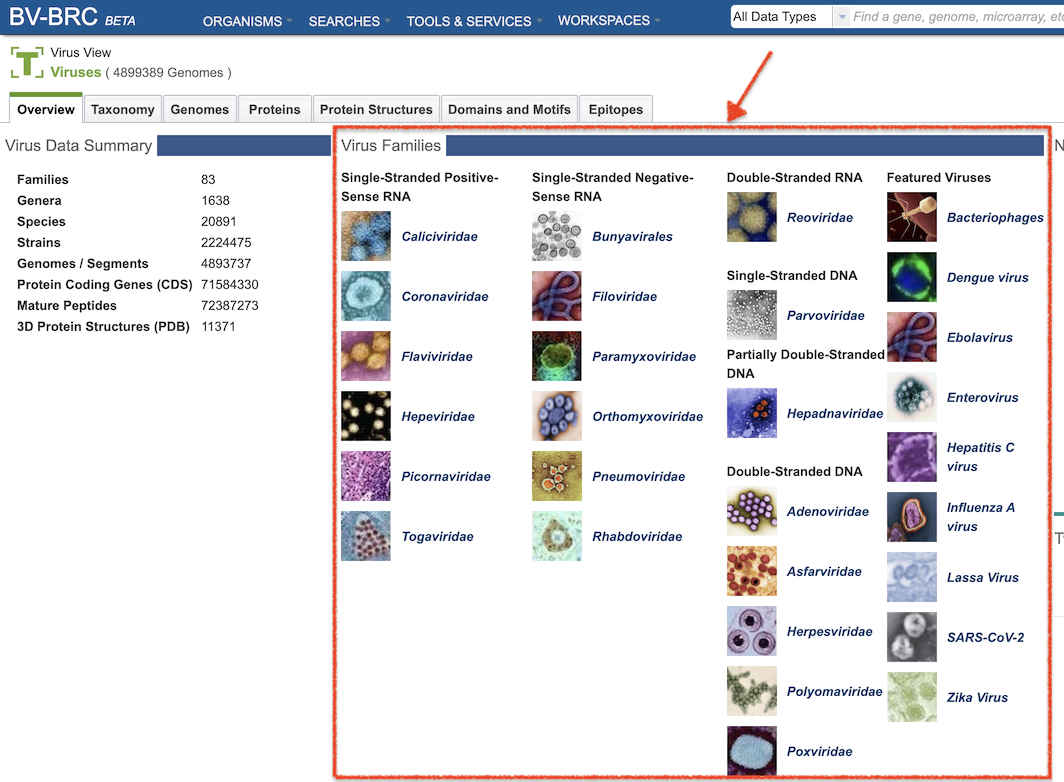
The BV-BRC Advanced Searches are available from the SEARCHES top menu, as shown below.
BV-BRC Advanced Searches
Advanced Searches are available for each of the major data types in BV-BRC.
Clicking on one of the Advanced Searches (e.g., “Strains”) opens a search form with drop down box options for primary metadata values and other special criteria.
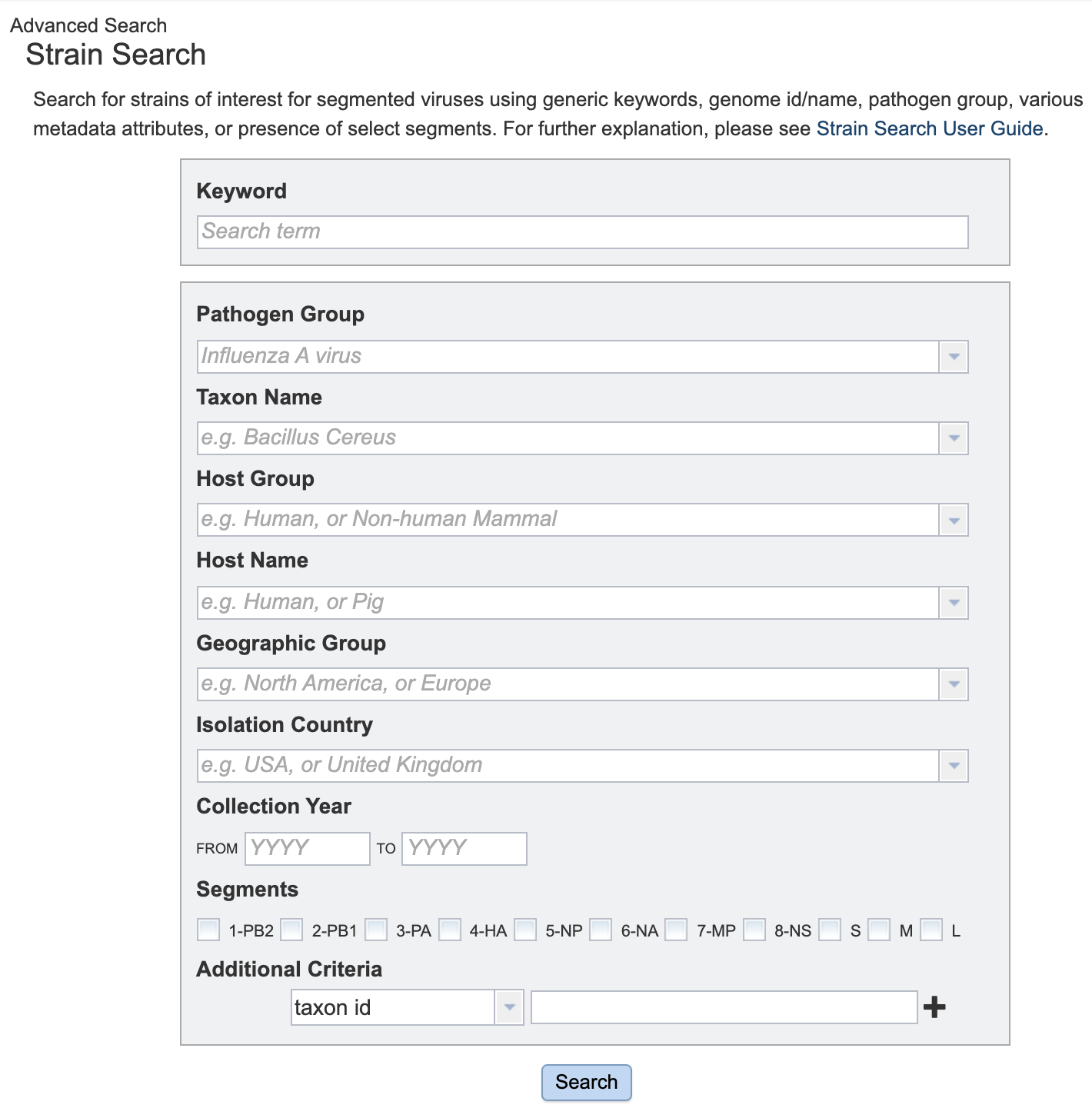
Detailed instructions for using the Advanced Searches are available from the BV-BRC Advanced Searches Quick Reference Guide.
The table below provides a mapping of the IRD/ViPR Search Tools to corresponding BV-BRC Advanced Searches.
Mapping of IRD/ViPR Search Tools to BV-BRC Advanced Searches
Analysis Tools¶
Similar to IRD and ViPR, BV-BRC provides tools and visualizations to enable researchers to perform in-depth analyses and exploration of their own data and in combination with BV-BRC data. BV-BRC integrates the most used analysis tools In IRD/ViPR (available from the IRD/ViPR Analysis Tools page, below).
IRD/ViPR Analysis Tools
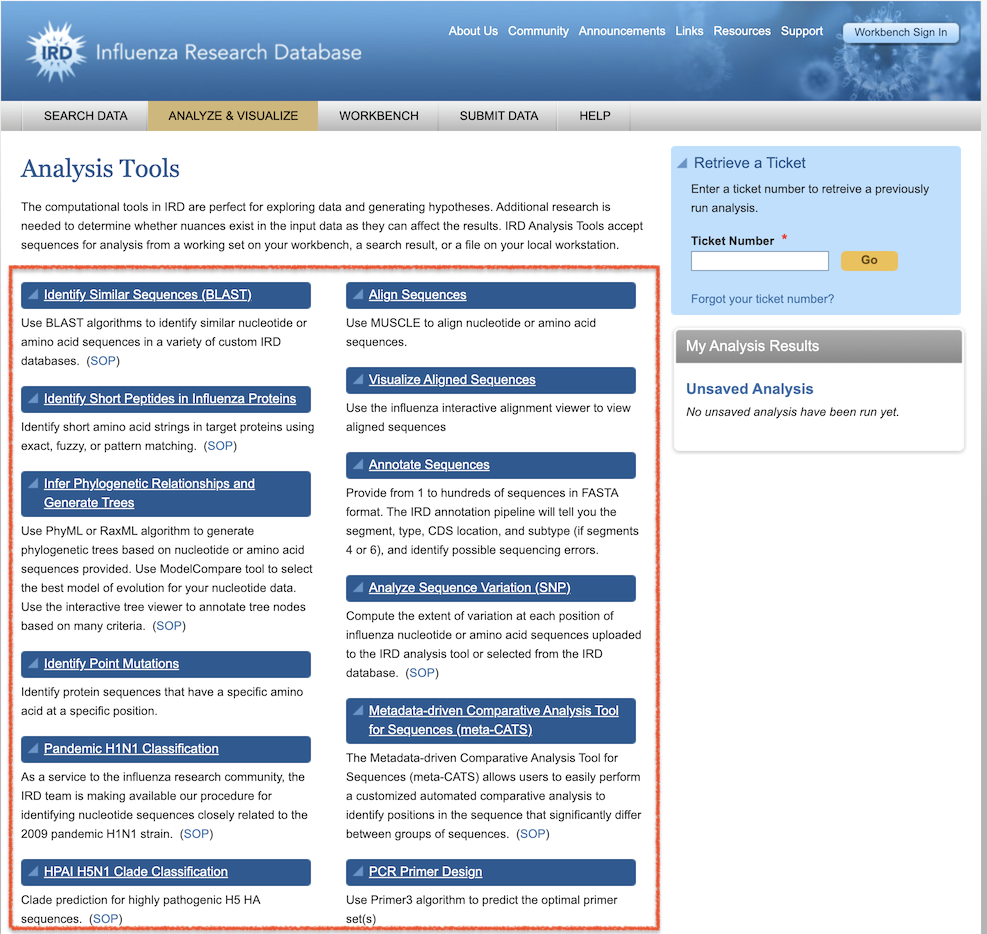
In BV-BRC, analysis tools are available from the TOOLS & SERVICES top menu, as shown below. A list of all tools with descriptions is available from the BV-BRC Tools & Services page
BV-BRC Tools & Services
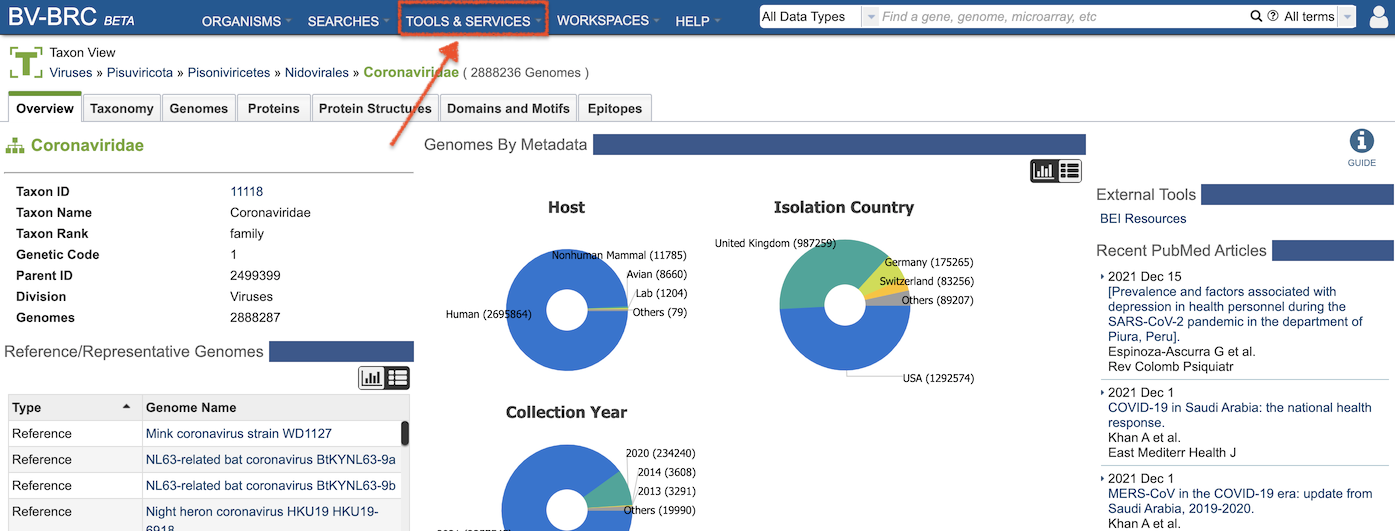
In the BV-BRC TOOLS & SERVICES menu, viral analysis tools are shown alongside bacterial analysis tools. Where practical, the two have been merged into one tool, with appropriate settings and logic to perform the correct type of computation based on the the organism type (e.g., Genome Annotation). The figure below highlights tools that have been added or updated to support viral data.

Clicking on one of the tools (e.g., “Annotation”) opens an input form that allows users to upload their own data, set parameters, and specify a location in the Workspace to save the results. Links to Quick Reference Guides and Tutorials are available on every input form. Quick Reference Guides provide short descriptions of each feature of the tool. Tutorials provide detailed, step-by-step instructions for using the tool.
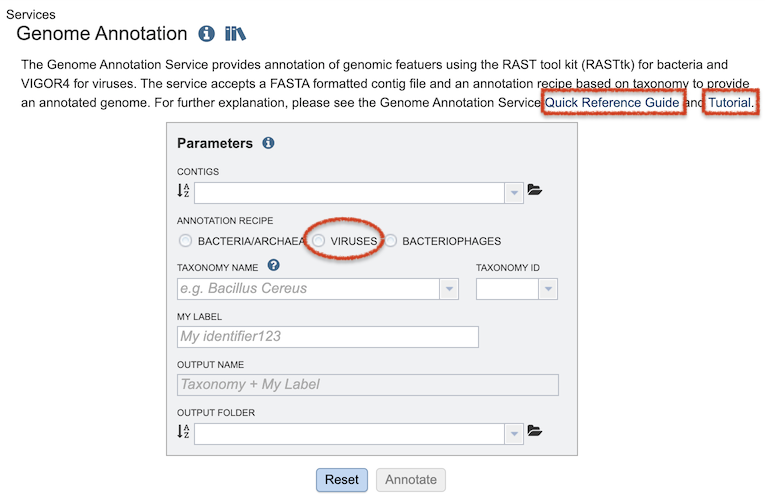
Most analysis tools in BV-BRC are implemented as computational “services” that use high-performance computing (HPC) resources to perform the analysis on the backend. When an analysis tool is started, it creates a “job” that runs on the HPC resources. The job processing status (Queued, Running, Complete, Failed) and results are listed on the user’s Job Status page, which is displayed by clicking the small Jobs status monitor on the bottom right of each page.
The Tools & Services Quick Reference Guide provides an overview available tools. The table below provides a mapping of the IRD/ViPR Search Tools to corresponding BV-BRC Advanced Searches.
| IRD/ViPR | BV-BRC | Documentation |
|---|---|---|
|
Identify Similar Sequences (BLAST) Identify Short Peptides in Proteins |
BLAST (Homology) |
Quick Reference Tutorial |
| Generate Phylogenetic Tree | Phylogenetic Tree (Gene Tree) |
Quick Reference Tutorial |
|
Align Sequences (MSA) Analyze Sequence Variation Visualize Aligned Sequences |
MSA and SNP Analysis |
Quick Reference Tutorial |
| Genome Annotation (VIGOR4) | Genome Annotation |
Quick Reference Tutorial |
| Metadata-driven Comparative Analysis Tool (Meta-CATS) | Metadata-driven Comparative Analysis Tool (Meta-CATS) |
Quick Reference Tutorial |
| Polymerase Chain Reaction (PCR) Primer Design | Primer Design |
Quick Reference Tutorial |
| Influenza H5, H3, H1 clade, rotavirus genotype, and flavivirus serotype classification | Subspecies Classification | Coming soon. |
Workbench¶
Similar to the IRD/ViPR Workbench (shown below), BV-BRC provides a private Workspace, where users can upload their own data, perform analyses with BV-BRC Tools, compare their data with other data in BV-BRC, and share their data via shared and public workspaces.
IRD/ViPR Workbench
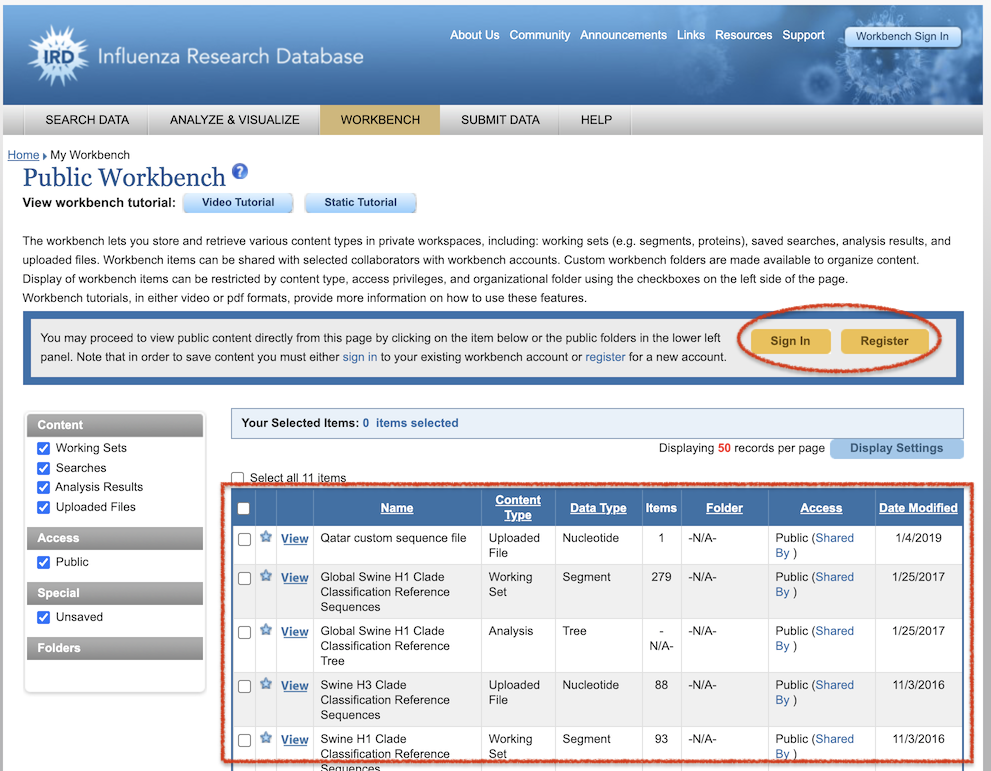
As with the IRD/ViPR Workbench, users will need to sign in (top right of page) to access their BV-BRC Workspace. IRD/ViPR users can use the same login ID and password as they do for IRD/ViPR. At present, we do not have the capability to transfer over data from IRD/ViPR Workbenches to the BV-BRC Workspace.
BV-BRC Workspace Login
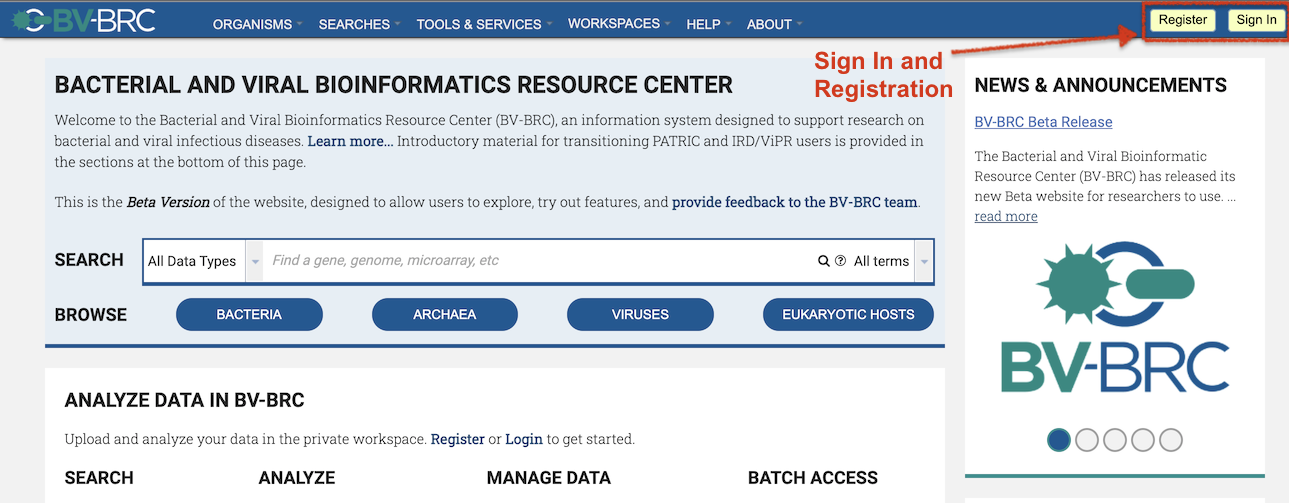
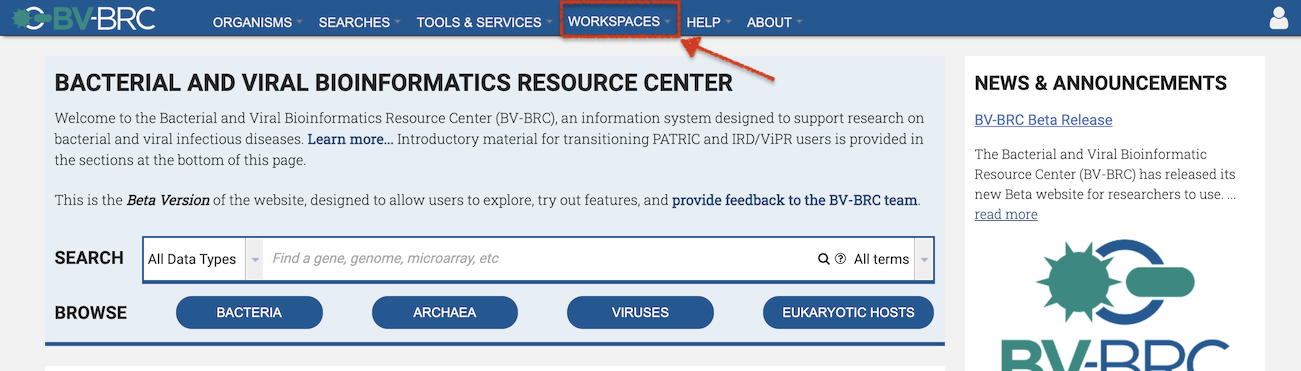
Once signed in, users can access their private and shared workspaces from the WORKSPACES top menu.
Note that, as of November 1, 2022, all IRD/ViPR workbench data and files were copied into the user’s corresponding workspace in BV-BRC. The data are organized into three folders: Working Sets, Uploaded Files, and Analysis Results. You can access this workspace with the same login credentials that you have for IRD/ViPR. There will be a README.txt file in the workspace that provides additional details.
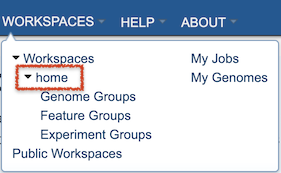
Clicking on the “Home” workspace opens the user’s private workspace at the top level. The Workspace contains folders to facilitate organization of files. Users can upload files and create new folders. Move, copy, delete, and other controls are also available.
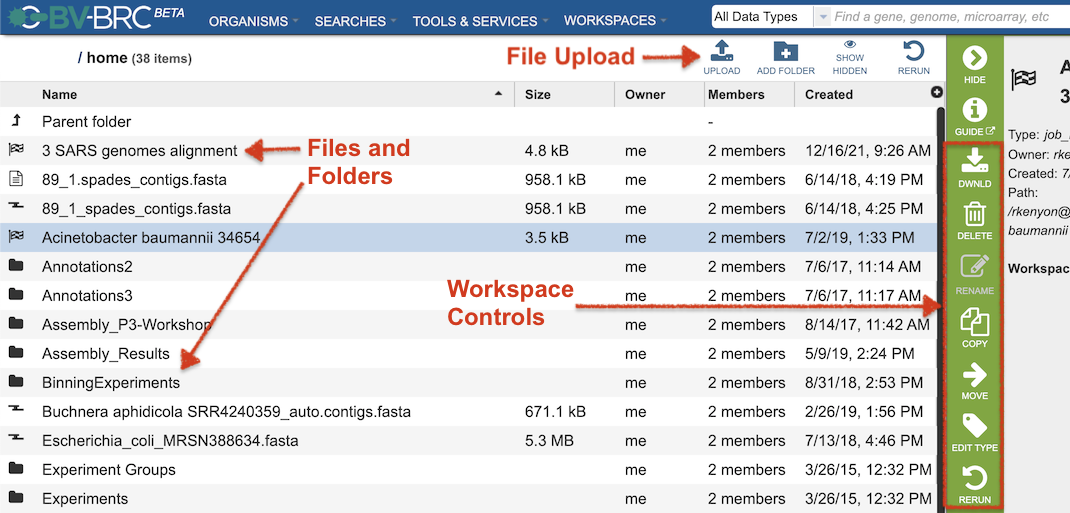
Also, users can create Groups of genomes, features (genes), and other data types, similar to IRD/ViPR “Working Sets.”
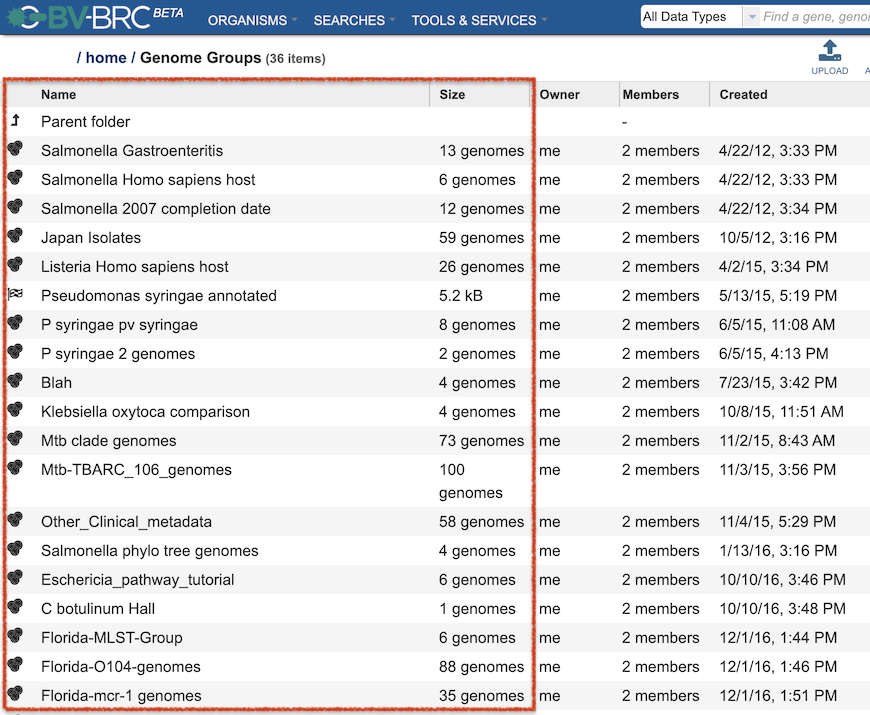
Detailed information on using the BV-BRC Workspace can be found in the Workspace, Private Data, Groups, Jobs Quick Reference Guides.